-
Discovery Science
Cracking the code — how the Human Phenotype Ontology advances genetic discovery
There are more than 7,500 known genetic disorders in humans, with new ones being added to the list on a regular basis. Yet pinpointing the single genetic change responsible for a constellation of characteristics can be challenging. Researchers at Mayo Clinic and elsewhere have tapped a resource known as the Human Phenotype Ontology (HPO) to help them identify previously unknown genetic diseases and diagnose the people who have them.
The HPO provides a standardized vocabulary — or ontology — for describing the many manifestations — or phenotypes — that arise from a particular genetic disorder. It includes more than 13,000 terms describing various abnormalities, such as "atrial septal defect" (a hole in the wall of the heart) or "arachnodactyly" (long, spider-like fingers). The ontology contains more than 156,000 annotations linking these terms to hereditary diseases.
"It allows you to structure a patient's phenotypes in a way that shows the relationships between them," says Eric Klee, Ph.D., a clinical genomics researcher at Mayo Clinic.
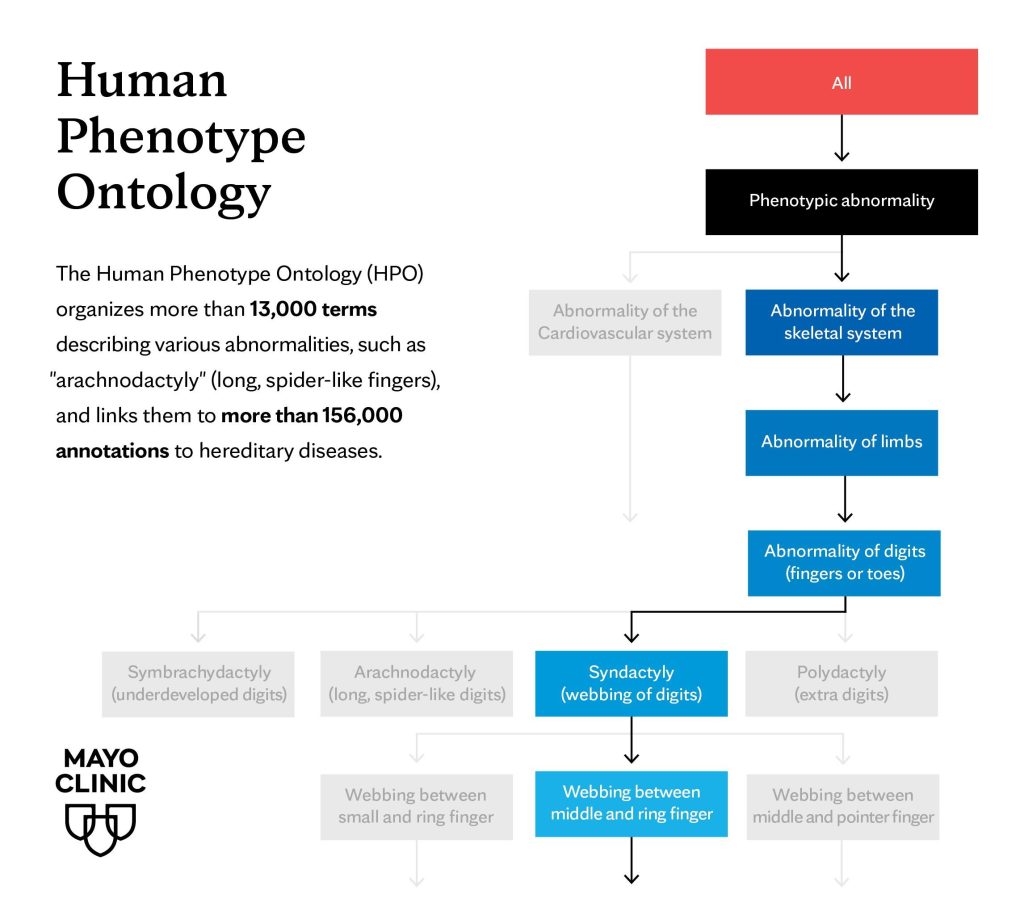
Specifically, the HPO organizes phenotypes into a hierarchical tree, with general categories at the top and more specific traits branching out below. For example, "Abnormal limb morphology" might be a broad category, with specific conditions like "arachnodactyly," "polydactyly" (extra fingers or toes) and "syndactyly" (webbed fingers or toes) as more detailed entries.
Dr. Klee says that this framework allows researchers to associate each phenotype with an exact number, such as arachnodactyly (HP0001166), effectively turning the description in a clinician's notes into a "computable format" that machines can help parse out.
"This helps us conduct what we call genotype-phenotype studies," he says. "In order to harness the power of what we see in the genetic databases, what we find in the scientific literature, and what we generate from sequencing patient DNA, we really do need computational tools."
Dr. Klee says that the HPO could be used not only to help interpret the results of genomic sequencing but also to help clinicians determine which patients might benefit from having their genomes sequenced in the first place.
"Patients with ultra-rare genetic disorders typically present with a nontypical cluster of symptoms, causing them to go on a diagnostic odyssey where they bounce around from provider to provider and from specialist to specialist before finally getting a diagnosis," he says.
Dr. Klee and his colleagues are testing whether clinicians can pair HPO terms with computational tools such as natural language processing and machine learning to help identify patients with these rare diseases earlier, when they are first seen by primary care physicians.
"Though individually rare, the collection of rare diseases is pretty common, affecting 5-8% of the population," he says. "If we could use these new tools to help identify these patients, that could be very valuable."







